Introduction
Purpose
OSA is a protocol for scientific data archives. It standardizes how data flows through deposition, validation, curation, and publication: the same workflow used by successful platforms like the Protein Data Bank, UniProt, and EMBL-EBI services.
The protocol separates infrastructure from domain logic. It defines the universal mechanics (how submissions move through stages, how validators report metrics, how nodes federate for discovery) while letting each scientific community plug in their own schemas and validation rules. A materials science archive and a genomics repository run the same protocol; only the domain-specific validators differ.
This means any scientific community can deploy professional-grade data infrastructure without rebuilding the data management logic from scratch.
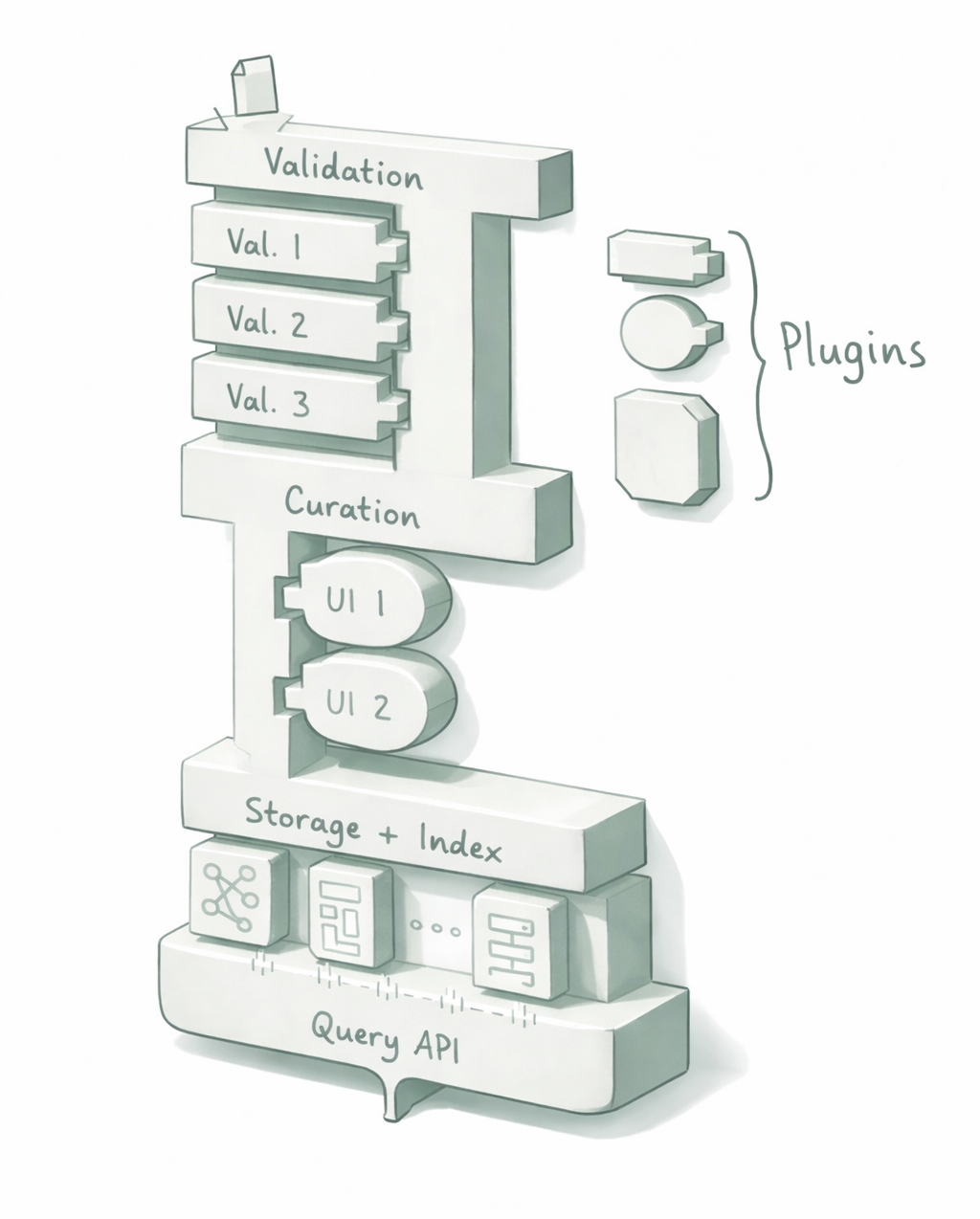
Motivation
Scientific data infrastructure follows a common pattern. Successful platforms like the Protein Data Bank (PDB), UniProt, Gene Expression Omnibus (GEO), and services at EMBL-EBI all implement the same core workflow: structured deposition, automated validation, expert curation, and programmatic access.
Despite this shared pattern, each new scientific domain rebuilds this infrastructure from scratch. This fragmentation results in:
- Duplicated effort: Generic pipeline logic is reimplemented rather than reused
- Inconsistent quality: Ad-hoc validation rules vary wildly across repositories
- Poor interoperability: Custom APIs prevent unified tooling and federated access
- High barriers: Emerging fields lack resources to build “PDB-quality” infrastructure
The OSA protocol addresses this by separating infrastructure from domain logic. The protocol defines how data flows through deposition, validation, curation, and publication (the universal “shape” of scientific data management). Domain-specific rules (what makes a protein structure valid vs. a materials dataset) are injected as pluggable components, not hard-coded into the platform.
This separation enables:
- Reusable implementations: A reference implementation serves as “PDB-in-a-box” for any domain
- Shared tooling: A dataset browser built for biology works immediately in physics
- Quality transparency: Machine-readable Traits let consumers filter by verified properties, not just file types
- Institutional flexibility: Existing platforms can expose data via OSA adapters without migration
By standardizing the infrastructure layer, OSA allows scientific domains to focus resources on what matters: their specific validation rules, curation workflows, and discovery interfaces, not rebuilding basic data pipelines.
Scope
This specification is implementation-agnostic. It defines what must be observable over the network, not how systems are organized internally.
In Scope
- Protocol resources: Structure and semantics of Depositions, Records, Vocabularies, and Tools
- State machines: Valid state transitions and invariants
- API contracts: HTTP endpoints and request/response formats
- Execution contracts: OCI container interfaces for Validators and Curation Tools
- Discovery protocols: Node discovery and federation
Out of Scope
- Internal architecture: How implementations organize code, services, or databases
- Storage mechanisms: Whether data is stored on S3, local disk, or elsewhere
- Authentication: Authentication mechanisms will be specified in a future OEP
- Performance characteristics: Caching strategies, indexing approaches, etc.
- Domain-specific validators: The protocol defines how to package validators, not what they check
Conformance Language
The key words “MUST”, “MUST NOT”, “REQUIRED”, “SHALL”, “SHALL NOT”, “SHOULD”, “SHOULD NOT”, “RECOMMENDED”, “MAY”, and “OPTIONAL” in this document are to be interpreted as described in RFC 2119.
Audience
This specification is intended for:
- Implementers building OSA-compliant ArchiveNodes or Index Nodes
- Tool developers creating Validators or Curation Tools
- Client developers building applications that interact with OSA nodes
- Governance bodies establishing policies for OSA ecosystems
Architecture
Actors
The OSA protocol defines five types of actors:
ArchiveNode : A service that accepts data submissions, orchestrates validation and curation, and publishes immutable Records. The primary write-path actor.
Index Node : A service that computes derived attributes about datasets, stores the results with provenance, and exposes a queryable, federated API. Index Nodes can process data from ArchiveNodes or external sources (e.g., GEO, SRA). They do not store raw data, only computed attribute values.
Validator
: An OCI container that computes vocabulary attributes from datasets. Validators emit metric values (e.g., mapped-reads-percent: 78.4), not pass/fail verdicts.
Curation Tool : An interactive OCI container that provides web-based interfaces for human reviewers to inspect and modify datasets.
Client : Any application (web app, CLI, library) that interacts with ArchiveNodes or Index Nodes to submit or retrieve data.
High-Level Flow
A typical dataset journey through OSA involves:
- Submission: A Client creates a Deposition on an ArchiveNode, uploads files, and submits for review.
- Validation: The ArchiveNode executes Validators to compute quality attributes for the data.
- Curation: A human curator uses a Curation Tool (proxied by the ArchiveNode) to review, annotate, or fix issues.
- Publication: Once approved, the ArchiveNode creates an immutable Record with a permanent identifier.
- Indexing: Index Nodes compute additional attributes and enable federated search across multiple sources.
┌────────┐
│ Client │
└────┬───┘
│ (1) Create Deposition
▼
┌─────────────┐
│ ArchiveNode │◄──(2) Run Validators
└─────────────┘
│
│ (3) Proxy Curation Tool
▼
┌──────────────┐
│ Curator │
└──────────────┘
│
│ (4) Approve → Create Record
▼
┌──────────┐ (5) Index
│ Index Node │◄─────────────┐
└──────────┘ │
▲ │
│ │
└────────────────────┘
(6) Search/DiscoverKey Concepts
Separation of Primary and Derived Data : ArchiveNodes hold primary data and manage the submission lifecycle. Index Nodes compute and index derived attributes without storing raw data. This separation enables specialisation: institutions run archives, quality-focused groups run indexes.
Metrics, Not Verdicts
: Validators emit numeric measurements (e.g., mapped-reads-percent: 78.4), not pass/fail judgments. Users filter datasets at query time with their own thresholds. This enables flexible, use-case-specific quality filtering.
Pluggable Domain Logic : Instead of hard-coding validation rules or curation interfaces, OSA uses OCI containers that can be developed, versioned, and shared independently.
Federation via Gossip : Index Nodes discover each other and share computed attributes through a heartbeat gossip protocol. No central coordination required.
Provenance by Default : Every computed attribute is tagged with which validator produced it, which node ran it, and when. Users can verify claims or filter by trusted sources.
Terminology
Resources
Deposition : A mutable, in-progress dataset submission. The primary resource on the write path. Transitions through states (DRAFT, SUBMITTED, UNDER_REVIEW, APPROVED) before becoming a Record.
Record : An immutable, versioned, published dataset. The primary resource on the read path. Created from an approved Deposition.
Vocabulary
: A collection of named attributes with defined types and semantics, inspired by OpenTelemetry’s semantic conventions. Vocabularies define attributes, they do not imply who computes them. Attributes are referenced using the vocab-srn#attribute-name syntax (e.g., urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent).
Attributed Value : A measurement paired with its attribute reference and provenance. Validators emit attributed values. Each value records which validator computed it, which node ran it, and when.
Trait
: A named, saved query over vocabulary attributes. Traits are convenience abstractions for common filter patterns (e.g., “high-quality-rnaseq” = mapped-reads-percent >= 70 AND duplicate-percent <= 30). Traits are evaluated at query time, not computed by validators.
Executables
Validator : An OCI container that computes vocabulary attributes from datasets. Validators declare which attributes they emit and produce attributed values with typed measurements. They do not emit pass/fail verdicts.
Curation Tool : An OCI container that exposes a web interface for human inspection and modification of Depositions.
CurationToolRef : A resource describing a Curation Tool: its OCI image, exposed port, and capabilities (read-only vs read-write).
Infrastructure
Structured Resource Name (SRN)
: A URN-based, globally unique, location-independent identifier for any OSA resource (e.g., urn:osa:data.example.org:rec:abc123@v1). The domain component enables DNS-based discovery.
Node Document
: A JSON file at /.well-known/osa-node.json that advertises a node’s capabilities and API endpoint. Enables decentralized discovery.
Gossip : The protocol by which Index Nodes discover each other and share information about which nodes compute which attributes. Operates via periodic heartbeats to known peers.
Resources
This section specifies what resources exist in the OSA protocol and what properties they MUST have. Implementations MAY add additional properties but MUST include all required fields.
Deposition
A Deposition represents a dataset in progress. It is mutable until submitted for validation.
Required Properties
| Property | Type | Description |
|---|---|---|
srn | SRN | Unique identifier (type: dep) |
status | string | Current state: DRAFT, SUBMITTED, UNDER_REVIEW, or APPROVED |
metadata | object | User-provided descriptive metadata |
files | array | List of file objects (see below) |
created_at | ISO 8601 datetime | Timestamp of creation |
updated_at | ISO 8601 datetime | Timestamp of last modification |
File Object Structure
Each entry in files MUST include:
| Property | Type | Description |
|---|---|---|
name | string | Filename |
size | integer | Size in bytes |
checksum | string | SHA-256 hash (hex-encoded) |
uploaded_at | ISO 8601 datetime | Upload timestamp |
Optional Properties
Implementations MAY include:
validation_runs: Array of ValidationRun objectscurator_id: User ID of assigned curator (when in UNDER_REVIEW state)submitted_at: Timestamp when status changed to SUBMITTED
Record
A Record represents an immutable, published dataset. It is created from an approved Deposition.
Required Properties
| Property | Type | Description |
|---|---|---|
srn | SRN | Unique identifier (type: rec) with version (e.g., @v1) |
status | string | Current state: PUBLIC, EMBARGOED, or WITHDRAWN |
metadata | object | Final descriptive metadata |
files | array | List of published file objects (same structure as Deposition files) |
provenance | object | Origin information (see below) |
published_at | ISO 8601 datetime | Timestamp of publication |
Provenance Object Structure
| Property | Type | Description |
|---|---|---|
source_deposition | SRN | The Deposition this Record was created from |
approved_by | string | User ID of the approving curator |
approved_at | ISO 8601 datetime | Approval timestamp |
attributes | array | List of attributed values computed at time of approval |
Attributed Value Object
Each entry in the attributes array represents a computed measurement:
| Property | Type | Description |
|---|---|---|
attribute | string | Full attribute reference (vocab-srn#attribute-name) |
value | any | The computed value (type matches vocabulary definition) |
validator | SRN | The Validator that computed this value |
computed_at | ISO 8601 datetime | When the value was computed |
The attributes field enables filtering and discovery based on computed quality metrics. Users can query for datasets matching their own thresholds (e.g., “mapped-reads-percent > 70”) rather than relying on fixed pass/fail verdicts.
Record Versioning
Records are immutable. If changes are needed, a new version MUST be created with an incremented version number (e.g., @v1 → @v2). The new version MUST reference the previous version in its provenance.
Vocabulary
A Vocabulary defines a set of named attributes with standardized semantics, inspired by OpenTelemetry Semantic Conventions. Vocabularies define attributes, they do not imply who computes them. Any node can compute values for any vocabulary’s attributes.
Attribute Reference Syntax
Attributes are referenced using the vocab-srn#attribute-name syntax:
urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent
└─────────────────────────────┘ └──────────────────┘
vocabulary SRN attribute nameThis syntax:
- Is globally unique (vocabulary SRN ensures uniqueness)
- Supports federation (domain in SRN enables DNS-based discovery)
- Keeps attributes scoped to their vocabulary (the vocabulary is the trust/curation boundary)
Required Properties
| Property | Type | Description |
|---|---|---|
srn | SRN | Unique identifier (type: vocab) |
title | string | Human-readable name (e.g., “RNA-seq Quality Metrics”) |
description | string | Purpose and scope of this vocabulary |
attributes | array | List of attribute definitions (see below) |
Attribute Definition Object
| Property | Type | Description |
|---|---|---|
name | string | Attribute name within this vocabulary (e.g., mapped-reads-percent) |
type | string | Data type: string, int, float, boolean, datetime, enum |
description | string | What this attribute represents |
unit | string | Unit of measurement (optional, e.g., percent, kelvin, bytes) |
range | array | Valid range for numeric types (optional, e.g., [0, 100]) |
Example Vocabulary
{
"srn": "urn:osa:osa.org:vocab:rnaseq@1",
"title": "RNA-seq Quality Metrics",
"description": "Quality metrics for RNA sequencing data",
"attributes": [
{
"name": "mapped-reads-percent",
"type": "float",
"unit": "percent",
"range": [0, 100],
"description": "Percentage of reads aligned to reference genome"
},
{
"name": "duplicate-percent",
"type": "float",
"unit": "percent",
"range": [0, 100],
"description": "Percentage of PCR/optical duplicate reads"
},
{
"name": "rrna-contamination-percent",
"type": "float",
"unit": "percent",
"range": [0, 100],
"description": "Percentage of reads mapping to ribosomal RNA"
},
{
"name": "read-count",
"type": "int",
"description": "Total number of reads in the dataset"
}
]
}Attributes from this vocabulary would be referenced as:
urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percenturn:osa:osa.org:vocab:rnaseq@1#duplicate-percent
Trait
A Trait is a named, saved query over vocabulary attributes. Traits provide a convenient shorthand for common filter patterns.
Required Properties
| Property | Type | Description |
|---|---|---|
srn | SRN | Unique identifier (type: trait) |
title | string | Human-readable name (e.g., “High Quality RNA-seq”) |
description | string | What this trait represents |
query | object | Attribute conditions (see below) |
Query Object
The query object maps attribute references to conditions:
{
"srn": "urn:osa:geo-index.org:trait:high-quality-rnaseq@1",
"title": "High Quality RNA-seq",
"description": "RNA-seq datasets with good mapping and low duplication",
"query": {
"urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent": { "gte": 70 },
"urn:osa:osa.org:vocab:rnaseq@1#duplicate-percent": { "lte": 30 }
}
}Supported operators: eq, neq, gt, gte, lt, lte, in, exists.
Traits vs Direct Queries
Users can always query attribute values directly without using Traits. Traits are optional UX conveniences that communities may publish as shared standards.
CurationToolRef
A CurationToolRef describes an interactive review tool.
Required Properties
| Property | Type | Description |
|---|---|---|
srn | SRN | Unique identifier |
title | string | Human-readable name (e.g., “3D Molecule Viewer”) |
image | string | OCI image reference (e.g., docker.io/osa/ngl-viewer:1.2.0) |
default_port | integer | Port the container’s web server listens on |
capabilities | array | List of strings: read-only and/or read-write |
Identifiers
All resources in the OSA protocol MUST be addressable by a Structured Resource Name (SRN).
SRN Grammar
SRNs MUST follow this URN-based grammar:
urn:osa:{node-id}:{type}:{local-id}[@{version}]Components
urn:osa
: The fixed scheme prefix. All OSA identifiers begin with this.
{node-id}
: The globally unique identifier of the originating node, typically a domain name (e.g., osa.org, imperial-mat-sci.ac.uk). Node IDs MUST be DNS-safe.
{type}
: A short string identifying the resource type:
dep: Depositionrec: Recordvocab: Vocabularyschema: Schema definitiontrait: Traitval: Validatortool: Curation Tool
{local-id}
: A node-unique, opaque identifier. Implementations MAY use UUIDs, sequential IDs, or other schemes. Local IDs MUST be URL-safe.
@{version} (optional)
: A version identifier. REQUIRED for Records, OPTIONAL for other resources. Versions SHOULD follow Semantic Versioning 2.0 (e.g., @v1.0.0, @v2.3.1).
Examples
urn:osa:osa.org:vocab:materials-core@v1.0.0
urn:osa:osa.org:trait:high-quality-rnaseq@v1.0.0
urn:osa:imperial-mat-sci.ac.uk:dep:xyz789
urn:osa:imperial-mat-sci.ac.uk:rec:xyz789@v1
urn:osa:osa.org:trait:iso8601-datesVersioning Semantics
Records MUST include versions: Every Record SRN MUST include a version component (e.g., @v1). This enables immutable references.
Other resources MAY include versions: Schemas, Vocabularies, and Tools MAY use versions to track evolution while maintaining backwards compatibility.
Version resolution: When an SRN without a version is dereferenced, the node SHOULD return the latest version.
Lifecycles
Deposition Lifecycle
A Deposition progresses through the following states:
┌─────────┐ submit ┌───────────┐ ┌──────────────┐
│ DRAFT │─────────────▶│ SUBMITTED │─────────────▶│ UNDER_REVIEW │
└─────────┘ └───────────┘ (curator └──────────────┘
▲ claims) │
│ │
│ ▼
│ ┌──────────┐
│ │ APPROVED │
│ └──────────┘
│ │
│ ▼
└──────────────────────────────────────────── [Record Created]
(request changes)State: DRAFT
Entry conditions: Automatically entered when a Deposition is created.
Permitted operations:
- Metadata MAY be modified
- Files MAY be uploaded or deleted
- Validators MAY be run (for pre-submission checks)
Transition to SUBMITTED: A Client MAY request transition to SUBMITTED. The ArchiveNode MUST validate that required metadata fields are present before allowing the transition.
State: SUBMITTED
Entry conditions: Depositor has indicated the submission is complete.
Observable requirements:
- The Deposition MUST be immutable to the original depositor
- Configured Validators MUST be executed
- ValidationRuns MUST be created and linked to the Deposition
Transition to UNDER_REVIEW: The ArchiveNode MAY transition to UNDER_REVIEW when validation is complete.
Transition to DRAFT: If validation fails, the ArchiveNode MAY allow a curator to request changes, returning the Deposition to DRAFT with feedback.
State: UNDER_REVIEW
Entry conditions: The Deposition is awaiting or undergoing human review.
Permitted operations:
- A curator MAY instantiate Curation Tools to inspect the data
- Curators MAY modify metadata or files (via Curation Tools) to fix issues
- The curator MAY add annotations or comments
Transition to APPROVED: The curator MAY approve the Deposition.
Transition to DRAFT: The curator MAY request changes from the depositor.
State: APPROVED
Entry conditions: A curator has approved the Deposition.
Observable requirement: The ArchiveNode MUST create a Record from the approved Deposition. This is a terminal state for the Deposition.
Validation Gate
Implementations MAY cache ValidationRuns and reuse them if the Deposition data has not changed. Implementations MUST re-run Validators if files or metadata have been modified since the last run.
Record Lifecycle
Records are immutable after creation. They have a simpler lifecycle:
State: PUBLIC
The Record is openly accessible. This is the default state for newly created Records.
State: WITHDRAWN
The Record has been retracted. The metadata remains visible (with a “WITHDRAWN” marker), but files are no longer accessible. Withdrawals MUST include a reason in the metadata.
ArchiveNode HTTP API
This section defines the HTTP API that conforming ArchiveNodes MUST implement.
General Conventions
Base URL: All endpoints are relative to the ArchiveNode’s base URL (e.g., https://archive.example.org/api/v1).
Authentication: Requests MUST include a Bearer token in the Authorization header:
Authorization: Bearer <token>Content Type: Request and response bodies MUST use application/json unless otherwise specified.
Error Responses: Errors MUST return appropriate HTTP status codes and a JSON body:
{
"error": "error_code",
"message": "Human-readable description"
}Deposition Endpoints
Create Deposition
POST /depositions
Creates a new Deposition in DRAFT state.
Request Body:
{
"metadata": {
"title": "My Dataset"
}
}Response (201 Created):
{
"srn": "urn:osa:example.org:dep:abc123",
"status": "DRAFT",
"metadata": {
"title": "My Dataset"
},
"files": [],
"created_at": "2024-01-15T10:30:00Z",
"updated_at": "2024-01-15T10:30:00Z"
}Get Deposition
GET /depositions/{id}
Retrieves a Deposition by its local ID.
Response (200 OK): Full Deposition object (as above).
Update Deposition Metadata
PATCH /depositions/{id}
Updates metadata fields. Only valid in DRAFT state (or UNDER_REVIEW if the requester is a curator).
Request Body:
{
"metadata": {
"title": "Crystal Structure of Protein X",
"authors": ["Alice", "Bob"]
}
}Response (200 OK): Updated Deposition object.
Upload File
POST /depositions/{id}/files
Uploads a file to the Deposition.
Request: multipart/form-data with a file field.
Response (201 Created):
{
"name": "data.cif",
"size": 1048576,
"checksum": "sha256:abcdef123456...",
"uploaded_at": "2024-01-15T10:35:00Z"
}Delete File
DELETE /depositions/{id}/files/{filename}
Removes a file from the Deposition. Only valid in DRAFT state.
Response (204 No Content)
Submit for Review
POST /depositions/{id}/actions/submit
Transitions the Deposition to SUBMITTED state, triggering validation.
Response (200 OK):
{
"status": "SUBMITTED",
"message": "Validation in progress"
}List Validation Runs
GET /depositions/{id}/validations
Returns all ValidationRuns for this Deposition.
Response (200 OK):
{
"validations": [
{
"validator": "urn:osa:osa.org:val:rnaseq-qc@1.0",
"executed_at": "2024-01-15T10:36:00Z",
"attributes": [
{
"attribute": "urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent",
"value": 78.4
}
]
}
]
}Record Endpoints
List Records
GET /records
Returns paginated list of public Records.
Query Parameters:
page: Page number (default: 1)per_page: Results per page (default: 20, max: 100)
Response (200 OK):
{
"records": [
{
"srn": "urn:osa:example-archive:rec:xyz789@v1",
"status": "PUBLIC",
"metadata": { "title": "..." },
"published_at": "2024-01-15T12:00:00Z"
}
],
"pagination": {
"page": 1,
"per_page": 20,
"total": 150
}
}Get Record
GET /records/{id}
Retrieves a specific Record by local ID. If no version is specified, returns the latest version.
GET /records/{id}@{version}
Retrieves a specific version of a Record.
Response (200 OK): Full Record object.
Download Record File
GET /records/{id}/files/{filename}
Downloads a file from a Record.
Response (200 OK): File contents (with appropriate Content-Type and Content-Disposition headers).
Curation Endpoints
List Available Tools
GET /depositions/{id}/tools
Returns Curation Tools available for this Deposition.
Response (200 OK):
{
"tools": [
{
"srn": "urn:osa:osa.org:tool:ngl-viewer@1.2.0",
"title": "3D Molecule Viewer",
"capabilities": ["read-only"]
}
]
}Start Curation Session
POST /depositions/{id}/sessions
Starts a Curation Tool session (launches OCI container and returns proxy endpoint).
Request Body:
{
"tool_srn": "urn:osa:osa.org:tool:ngl-viewer@1.2.0"
}Response (201 Created):
{
"session_id": "sess_abc123",
"status": "provisioning",
"proxy_endpoint": "/curation/sessions/sess_abc123",
"expires_at": "2024-01-15T14:00:00Z"
}Access Curation Tool
GET /curation/sessions/{session_id}/{path}
Proxies requests to the running Curation Tool container.
The ArchiveNode MUST:
- Verify the requester owns the session
- Forward requests to the container’s web server
- Rewrite paths according to
OSAP_BASE_PATH
Stop Curation Session
DELETE /curation/sessions/{session_id}
Terminates the Curation Tool container.
Response (204 No Content)
Index Node Protocol
An Index Node computes derived attributes about datasets, stores the results with provenance, and exposes a queryable, federated API. Index Nodes do not store raw data, only computed attribute values.
Responsibilities
A conforming Index Node MUST:
- Run validators against datasets to compute vocabulary attributes
- Store attributed values with full provenance (which validator, which node, when)
- Expose a Search API for querying by attribute conditions
- Participate in gossip to enable federated discovery
Index Nodes MAY:
- Index data from ArchiveNodes (OSA-native records)
- Index data from external sources (GEO, SRA, ArrayExpress) via source adapters
- Aggregate results from multiple data sources
Data Model
Index Nodes store attributed values, not raw data:
dataset_id: GSE12345
source: geo
attributes:
- attribute: urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent
value: 78.4
provenance:
validator: urn:osa:salmon.org:val:salmon@2.1
node: urn:osa:geo-index.org:node:main
computed_at: 2025-12-01T12:00:00Z
- attribute: urn:osa:osa.org:vocab:rnaseq@1#duplicate-percent
value: 12.3
provenance:
validator: urn:osa:picard.org:val:markdup@3.0
node: urn:osa:geo-index.org:node:main
computed_at: 2025-12-01T12:05:00ZSearch API
Search by Attribute Conditions
GET /search
Searches datasets by attribute value conditions.
Query Parameters:
q: Attribute condition(s) in formatattribute:op:value(repeatable)source: Filter by data source (e.g.,geo,osa)page: Page number (default: 1)per_page: Results per page (default: 20, max: 100)
Example:
GET /search
?q=urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent:gte:70
&q=urn:osa:osa.org:vocab:rnaseq@1#duplicate-percent:lte:30
&source=geo
&limit=100Response (200 OK):
{
"results": [
{
"dataset_id": "GSE12345",
"source": "geo",
"attributes": {
"urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent": {
"value": 78.4,
"provenance": {
"validator": "urn:osa:salmon.org:val:salmon@2.1",
"node": "urn:osa:geo-index.org:node:main",
"computed_at": "2025-12-01T12:00:00Z"
}
},
"urn:osa:osa.org:vocab:rnaseq@1#duplicate-percent": {
"value": 12.3,
"provenance": { ... }
}
}
}
],
"pagination": {
"page": 1,
"per_page": 20,
"total": 42
},
"federated_from": ["urn:osa:geo-index.org:node:main"]
}Search by Trait
GET /search?trait={trait-srn}
Convenience endpoint that expands a Trait to its underlying attribute conditions.
Example:
GET /search?trait=urn:osa:geo-index.org:trait:high-quality-rnaseq@1Equivalent to querying with the Trait’s query conditions directly.
Gossip Protocol
Index Nodes discover each other and share information about which nodes compute which attributes through a gossip protocol.
Heartbeat
Each Index Node periodically sends heartbeat messages to its known peers:
POST /gossip/heartbeat
{
"node_id": "urn:osa:inst-a.edu:node:main",
"computes": [
"urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent",
"urn:osa:osa.org:vocab:rnaseq@1#duplicate-percent"
],
"peers": [
"urn:osa:inst-b.org:node:metrics",
"urn:osa:geo-index.org:node:main"
],
"timestamp": "2025-12-07T10:00:00Z"
}The computes field lists which vocab#attr combinations this node can compute. The peers field shares known peers for network discovery.
Peer Discovery
When a node starts computing attributes from a new vocabulary, it SHOULD:
- Resolve the vocabulary author’s domain via DNS
- Fetch their Node Document at
/.well-known/osa-node.json - Add them as a gossip peer
This ensures vocabulary authors naturally become hubs for their vocabulary’s ecosystem.
Gossip Catalog
Each node maintains a local catalog of which nodes compute which attributes:
catalog:
urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent:
- node: urn:osa:inst-a.edu:node:main
last_seen: 2025-12-07T10:00:00Z
- node: urn:osa:geo-index.org:node:main
last_seen: 2025-12-06T15:00:00ZCatalog entries expire after a configurable TTL without a heartbeat refresh.
Federated Queries
When a node receives a query for an attribute it doesn’t compute locally:
- Check gossip catalog for nodes that compute the attribute
- Fan out the query to those nodes
- Merge results, preserving provenance
- Return unified response with
federated_fromfield
POST /federation/query
{
"attributes": [
{
"attribute": "urn:osa:inst-b.org:vocab:qc@1#rrna-percent",
"op": "lt",
"value": 5
}
],
"dataset_ids": ["GSE12345", "GSE67890"],
"source": "geo"
}Trust Model
Every attributed value includes full provenance. Users can:
- Filter by trusted nodes: “only values from nodes I trust”
- Filter by validator: “only values computed by validator X”
- Verify independently: re-run the validator themselves
Nodes MAY implement trust policies for accepting gossiped information (accept all, allowlist, require signatures).
Validator Contract
This section defines the execution contract for Validator OCI containers.
Purpose
Validators are headless, automated programs that compute vocabulary attributes from datasets. They run in sandboxed OCI containers and communicate via files. Validators emit metric values, not pass/fail verdicts, filtering is done at query time.
Validator Manifest
Each validator MUST include a manifest file at /osa/manifest.json within the container:
{
"srn": "urn:osa:salmon.org:val:salmon@2.1.0",
"name": "Salmon Quantification",
"description": "Computes RNA-seq alignment and quantification metrics",
"emits": [
"urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent",
"urn:osa:osa.org:vocab:rnaseq@1#duplicate-percent",
"urn:osa:osa.org:vocab:sequencing@1#read-count"
]
}The emits field declares which vocab#attr combinations this validator produces, enabling discovery and gossip.
Container Requirements
Packaging
Validators MUST be packaged as OCI-compliant container images (compatible with Docker, Podman, etc.).
Entrypoint
The container’s entrypoint MUST:
- Read input data from
$OSAP_IN - Compute vocabulary attributes
- Write results to
$OSAP_OUT/result.json - Exit with code 0 if computation completed (regardless of data quality)
Environment Variables
The host MUST inject:
| Variable | Description |
|---|---|
OSAP_IN | Path to input directory containing data files |
OSAP_OUT | Path to output directory (writable) |
Input Format
The $OSAP_IN directory contains:
files/: Data files to processmetadata.json: Dataset metadata (optional)config.json: Per-run configuration (optional)
Output Format
Validators MUST write $OSAP_OUT/result.json with computed attribute values:
{
"attributes": [
{
"attribute": "urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent",
"value": 78.4
},
{
"attribute": "urn:osa:osa.org:vocab:rnaseq@1#duplicate-percent",
"value": 12.3
},
{
"attribute": "urn:osa:osa.org:vocab:sequencing@1#read-count",
"value": 45000000
}
],
"logs": [
"Processed 45M reads in 12m34s",
"Reference: GRCh38"
]
}Required fields:
attributes: Array of computed attribute values
Optional fields:
logs: Array of human-readable diagnostic messageserrors: Array of error messages (computation still completed)
Example Validation Run
# Host prepares input
$ ls $OSAP_IN/files/
sample_R1.fastq.gz sample_R2.fastq.gz
# Host runs container
$ docker run --rm \
--network none \
-v /path/to/input:/input:ro \
-v /path/to/output:/output \
-e OSAP_IN=/input \
-e OSAP_OUT=/output \
salmon.org/osa/salmon:2.1.0
# Validator writes result
$ cat $OSAP_OUT/result.json
{
"attributes": [
{"attribute": "urn:osa:osa.org:vocab:rnaseq@1#mapped-reads-percent", "value": 78.4}
]
}Sandboxing
Nodes MUST run Validators with:
- No network access (no outbound connections)
- Read-only input (
$OSAP_INmounted read-only) - Limited resources (CPU/RAM limits enforced)
- Timeout (execution killed after limit, e.g., 30 minutes)
- Isolated filesystem (no access to host beyond input/output)
Error Handling
| Scenario | Behavior |
|---|---|
| Exit code non-zero | Mark as error, log stderr, no attributes stored |
No result.json written | Mark as error, “No result produced” |
| Timeout exceeded | Kill container, mark as error, “Timeout exceeded” |
result.json malformed | Mark as error, “Invalid output format” |
Errors are recorded with the dataset. Users can see “validator X failed on this dataset” without it blocking other validators.
Curation Tool Contract
This section defines the execution contract for Curation Tool OCI containers.
Purpose
Curation Tools are interactive web applications that allow human curators to inspect, annotate, and modify Depositions. Unlike Validators, they are long-running and expose HTTP interfaces.
Container Requirements
Packaging
Curation Tools MUST be packaged as OCI-compliant container images.
Web Server
The container MUST run a web server listening on the port specified in its CurationToolRef.default_port (e.g., 8080).
Environment Variables
The ArchiveNode MUST inject:
| Variable | Description |
|---|---|
OSAP_DEPOSITION_ID | The SRN of the Deposition being curated |
OSAP_API_URL | The internal URL of the ArchiveNode’s API (e.g., http://localhost:5000/api/v1) |
OSAP_API_TOKEN | A temporary Bearer token with write access to the Deposition |
OSAP_BASE_PATH | The public path prefix (e.g., /curation/sessions/sess_abc123) |
Base Path Handling
All assets (HTML, CSS, JS, images) MUST be served relative to $OSAP_BASE_PATH.
Example: If the tool serves a stylesheet at /style.css, and OSAP_BASE_PATH=/curation/sessions/sess_abc123, the client must access it at:
https://archive.example.org/curation/sessions/sess_abc123/style.cssMany web frameworks support base path configuration (e.g., Flask’s APPLICATION_ROOT, Express’s app.use(basePath, ...)).
Data Access
Reading Data
Curation Tools MAY access Deposition data via:
- API calls to
$OSAP_API_URLusing$OSAP_API_TOKEN - Mounted files (if the ArchiveNode mounts data at
/dataread-only)
Modifying Data
To modify the Deposition (e.g., fix metadata, delete files), the tool MUST:
- Make API calls to
$OSAP_API_URL(e.g.,PATCH /depositions/{id}) - Include
Authorization: Bearer $OSAP_API_TOKEN
Tools MUST NOT attempt to write state to the container’s local filesystem. Any data written locally will be lost when the session ends.
Security
Token Scope
The provided OSAP_API_TOKEN:
- MUST grant read access to the Deposition being curated
- MUST grant write access ONLY if the tool’s
capabilitiesincludesread-write - MUST expire when the curation session ends
- SHOULD be limited to the specific Deposition (not all Depositions)
Isolation
Curation Tool containers MUST be isolated:
- Network restrictions: Outbound internet access SHOULD be blocked unless explicitly required
- Ephemeral storage: Any data written to the container’s filesystem MUST be discarded when the session ends
- Resource limits: CPU/RAM limits SHOULD be enforced
Example Curation Session
# ArchiveNode starts container
$ docker run --rm -d \
-p 8080:8080 \
-e OSAP_DEPOSITION_ID=urn:osa:example:dep:abc123 \
-e OSAP_API_URL=http://host.docker.internal:5000/api/v1 \
-e OSAP_API_TOKEN=eyJhbGc... \
-e OSAP_BASE_PATH=/curation/sessions/sess_xyz \
myregistry.io/osa/molecule-viewer:1.2.0
# ArchiveNode proxies requests
# User visits: https://archive.example.org/curation/sessions/sess_xyz
# ArchiveNode forwards to: http://localhost:8080/ (with path rewriting)Client Requirements
A Client is any application that interacts with ArchiveNodes or Index Nodes. This includes web applications, command-line tools, libraries, and scripts.
Required Behaviors
Conforming Clients MUST:
-
Use SRNs for resource references: When referencing Depositions or Records, use full SRNs (not local IDs alone).
-
Authenticate via Bearer tokens: Include
Authorization: Bearer <token>in all requests to ArchiveNodes. -
Handle standard HTTP status codes:
401 Unauthorized→ Prompt for authentication403 Forbidden→ Insufficient permissions404 Not Found→ Resource does not exist422 Unprocessable Entity→ Validation errors (check response body for details)
-
Respect rate limits: If an ArchiveNode returns
429 Too Many Requests, back off exponentially.
Optional Behaviors
Clients MAY:
- Cache Record metadata locally (but SHOULD check
ETagorLast-Modifiedheaders) - Support multiple ArchiveNodes simultaneously
- Implement retry logic for transient failures (5xx errors)
Example: Submitting a Dataset
import requests
API_BASE = "https://archive.example.org/api/v1"
TOKEN = "your-bearer-token"
# 1. Create Deposition
resp = requests.post(
f"{API_BASE}/depositions",
headers={"Authorization": f"Bearer {TOKEN}"},
json={"metadata": {"title": "My Crystal Structure"}}
)
deposition = resp.json()
dep_id = deposition["srn"].split(":")[-1] # Extract local ID
# 2. Upload file
with open("data.cif", "rb") as f:
requests.post(
f"{API_BASE}/depositions/{dep_id}/files",
headers={"Authorization": f"Bearer {TOKEN}"},
files={"file": f}
)
# 3. Update metadata
requests.patch(
f"{API_BASE}/depositions/{dep_id}",
headers={"Authorization": f"Bearer {TOKEN}"},
json={"metadata": {"title": "My Crystal Structure"}}
)
# 4. Submit
requests.post(
f"{API_BASE}/depositions/{dep_id}/actions/submit",
headers={"Authorization": f"Bearer {TOKEN}"}
)Security & Privacy
Authentication
ArchiveNodes MUST support Bearer token authentication. Tokens SHOULD be obtained via an external OIDC-compatible identity provider.
ArchiveNodes MAY support additional authentication methods (e.g., API keys, mTLS) but MUST support Bearer tokens for interoperability.
Authorization
ArchiveNodes MUST enforce:
- Depositor isolation: Users can only read/modify Depositions they created (unless they have curator privileges)
- Curator privileges: Curators can view and modify any Deposition in UNDER_REVIEW state
- Public read access: Records in PUBLIC state SHOULD be readable without authentication
Node Identity
Each ArchiveNode MUST have a globally unique NodeID and MUST publish a Node Document at:
https://{domain}/.well-known/osa-node.jsonExample:
{
"node_id": "urn:osa:imperial-mat-sci.ac.uk:node:main",
"version": "1.0.0",
"api_base": "https://imperial-mat-sci.ac.uk/osa/api/v1",
"capabilities": ["archive", "index"],
"peers": [
"urn:osa:geo-index.org:node:main"
]
}Curation Tool Security
Proxy Authentication
The ArchiveNode’s proxy endpoint (/curation/sessions/{session_id}) MUST verify that the requesting user is the owner of the session.
Tool Isolation
Curation Tools MUST be isolated:
- Network restrictions: Outbound internet access SHOULD be blocked
- Ephemeral storage: Any data written to the container’s filesystem MUST be discarded when the session ends
- No persistent state: Tools MUST NOT maintain state between sessions
CSRF Protection
Because Curation Tools are proxied on the ArchiveNode’s domain, they share the same origin as the main application. ArchiveNodes MUST:
- Enforce
SameSite=Stricton session cookies - Validate CSRF tokens on all state-changing operations
- Use separate session tokens for curation (not the main user session)
Data Privacy
ArchiveNodes SHOULD:
- Encrypt data at rest and in transit (TLS 1.3+)
- Provide mechanisms for embargoing sensitive data
- Support metadata redaction for withdrawn Records
- Log all access to private Depositions for audit purposes
Conformance
Conformance Classes
ArchiveNode
A conforming ArchiveNode MUST:
- Implement all endpoints in ArchiveNode HTTP API
- Enforce the Deposition lifecycle (Lifecycles)
- Execute Validators according to Validator Contract
- Proxy Curation Tools according to Curation Tool Contract
- Support SRN resolution (Identifiers)
- Publish a Node Document at
/.well-known/osa-node.json
Index Node
A conforming Index Node MUST:
- Implement the Search API (Index Node Protocol)
- Index Records from at least one data source (ArchiveNode or external archive via Source Adapter)
- Store attributed values with full provenance (validator, node, timestamp)
- Support filtering by vocabulary attributes via query parameters
- Resolve SRNs (Identifiers)
- Return results in the specified JSON format
- Publish a Node Document at
/.well-known/osa-node.json - Participate in gossip protocol with at least one peer
Validator
A conforming Validator MUST:
- Be packaged as an OCI container
- Read input from
$OSAP_IN - Write
result.jsonto$OSAP_OUTcontaining anattributesarray of attributed values - Each attributed value MUST include
attribute(vocab#attr reference) andvalue - Exit with code 0
- Operate without network access
Curation Tool
A conforming Curation Tool MUST:
- Be packaged as an OCI container
- Run a web server on the specified port
- Serve assets relative to
$OSAP_BASE_PATH - Use
$OSAP_API_TOKENfor all state-changing operations - Not write persistent state to local disk
Client
A conforming Client MUST:
- Use SRNs for resource references
- Authenticate via Bearer tokens
- Handle standard HTTP status codes
Version Compatibility
This specification is version 0.0.1-alpha.
Breaking changes (as defined by Semantic Versioning 2.0) will increment the major version. Implementations SHOULD advertise their supported spec versions in the Node Document.
Extensibility
OSA Enhancement Proposals (OEPs)
Changes to this specification or standard contracts follow the OEP process. OEPs are published at https://oeps.opensciencearchive.org/.
Namespaced Extensions
Implementations MAY add custom fields to protocol resources using namespaced keys:
{
"metadata": {
"title": "My Dataset",
"x-institution-grant-id": "GRANT-12345",
"x-institution-internal-id": "abc-xyz-789"
}
}Extension keys MUST:
- Start with
x-followed by a unique namespace identifier - Use lowercase and hyphens (e.g.,
x-my-org-field-name) - Not conflict with standardized keys
Extensions MUST NOT:
- Change the semantics of required protocol fields
- Break interoperability (other implementations MUST be able to ignore unknown fields)